  |
Using the sign-up form
1) Accept cookies if asked.2) Click on the arrow icon in the upper right corner.
3) Choose "Sign up".
4) Provide your email. If you are worried about your privacy - please read our FAQ.
5) Create a strong password. Read our instruction about creating strong password. Please read our help article on passwords.
6) Confirm the chosen password and press "Sign up" button.
Here you go! You are now a registered user. Before using your account you have to activate it. You will receive a mail with the activation link.
Then do not forget to type in a new project ID in the project box (upper row) and press Enter. It should shine green. Then every time you login, your project(s) would be in the drop-down list. The user file lists (under "Altered gene sets" and "Functional gene sets") will be available if you press the respective icons in "File".
| How to begin? | ||
| Access, users, projects | ||
| Identification of driver genes | ||
| Exploration of molecular landscapes | ||
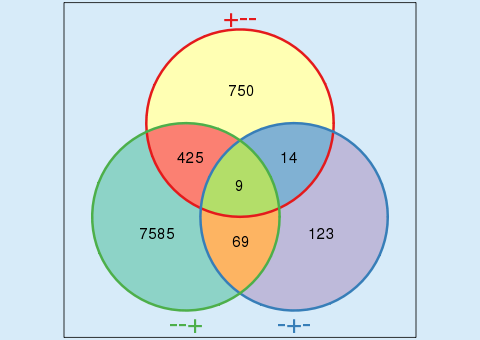
| Using Venn diagrams | ||
| Using stand-alone software |
Potentially, you can obtain much more useful output by running the perl script NEA.pl off-line.
NEA.pl is the script that executes network enrichment analysis at this web site.
Please note that while running analysis online, on this web-site, you can both look up the command line and download specific input and output files.
Watch the text and links that appear after the current analysis finished.
You can to use them either as a reference or as input to a modified analysis off-line. _tmpAGS, _tmpFGS, and _tmpNET contain your submitted gene list(s), the collection of functional gene sets, and the network.
R package NEArender on local site
perl script NEA.pl from Merid et al. (2014)
SomaticMutations.TCGA-2008
TAB-delimited file with results of a differential expression analysis, gene IDs followed by fold change, p, and FDR values in multiple contrasts:
P.matrix.NoModNodiff_wESC.DE.VENN.txt
This site best works with Google Chrome (v. 60 or later) and Mozilla Firefox (v. 57 or later).
Some functions do not work with IE 11 (opening uploaded user's files)..
The site also works with Apple Safari (v. 11.1, macOS Sierra v10.12.6), other versions of Safari browser can have compatibility issues.
Ashwini Jeggari, Zhanna Alekseenko, José Dias, Johan Ericson, and Andrey Alexeyenko EviNet: a web platform for network enrichment analysis with flexible definition of gene sets Nucleic Acids Res 2018 Jul 2;46(W1):W163-W170. doi: 10.1002/1878-0261.12350.
The method of network enrichment analysis (NEA):Andrey Alexeyenko, Woojoo Lee, Maria Pernemalm, Justin Guegan, Philippe Dessen, Vladimir Lazar, Janne Lehtiö and Yudi Pawitan Network enrichment analysis: extension of gene-set enrichment analysis to gene networks BMC Bioinformatics 2012 Sep 11;13:226 doi: 10.1186/1471-2105-13-226.
The first implementation of the fast NEA algorithm:Andrey Alexeyenko, Deena M. Wassenberg, Edward K. Lobenhofer, Jerry Yen, Erik L.L. Sonnhammer, Elwood Linney and Joel N. Meyer Dynamic zebrafish interactome reveals transcriptional mechanisms of dioxin toxicity PLoS One 2010 May 5;5(5):e10465 doi: 10.1371/journal.pone.0010465.
The back-end software:Ashwini Jeggari and Andrey Alexeyenko NEArender: an R package for functional interpretation of ‘omics’ data via network enrichment analysis BMC Bioinformatics 2017 Mar 23;18(Suppl 5):118. doi: 10.1186/s12859-017-1534-y.
